Research Thrust 2
Targeted delivery methods
Projects in Research Thrust 2 (RT2) will build the toolbox needed for targeting the delivery/removal of desired genetic features or vectors in the built environment as well as enabling functional modulation in an established microbial community.
RT2 researchers will use their knowledge of delivery systems and nanoparticle transport in complex environments to develop the requisite toolbox for microbiome engineering in the built environment. Initial projects will focus on limiting the spread of antimicrobial resistance (AMR) in built microbiomes as well as targeting the built environment water microbiome as there is a critical body of work linking the microbiome of premise plumbing to adverse effects in the built environment. Later efforts will transition to engineering solutions to limiting pathogens and bioaerosols as PreMiEr’s research matures.
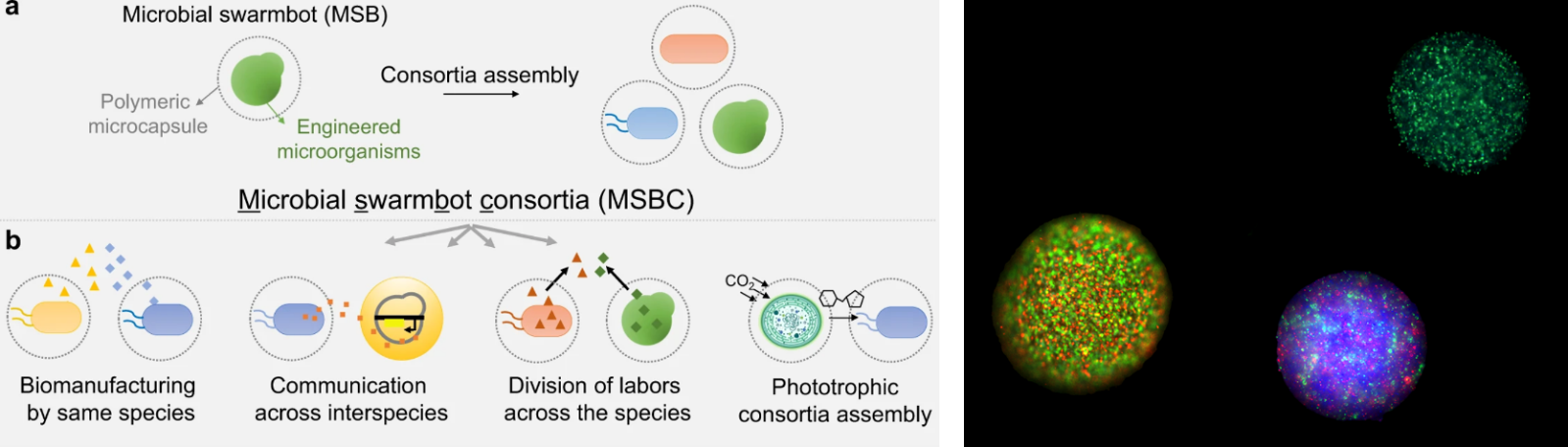
Left image from : Wang et al. 2022. Engineering consortia by polymeric microbial swarmbots. Nature Comm. 13: 3879.
Right image : Swarmbot delivery vehicles containing microorganisms. Photo courtesy of Lingchong You.
Currently funded projects
RT2-1: Developing engineering tools for plumbing-associated microbes
This project develops plasmid and phage systems for the genetic manipulation of microbes in the built environment. These tools will allow 1) upregulation/downregulation/knockout of individual genes within built environment microbes, and 2) delivery of genetic material to specific microbes in situ, paving the way for functional studies. As a representative and important “built environment” habitat, this project is focusing on premise plumbing, including sinks, drains, and toilets.
Collaborations
![]()
![]()
![]()
![]()
![]()

Nathan Crook
NC State, Project Lead

Kevin Garcia
NC State

Claudia Gunsch
Duke

Jennifer Kuzma
NC State

Robert Newman
N.C. A&T

Lingchong You
Duke

Yihui Zhou
NC State
RT2-2: Evaluation of biopolymeric microcapsules for the delivery of targeted microbes in premise plumbing systems
Traditionally, waterborne disease outbreaks have been prevented through centralized water treatment in utility water treatment plants. However, several CDC studies have shown that a significant number of waterborne disease outbreaks result from opportunistic pathogens that reside in premise plumbing environments as opposed to those associated with either water treatment plants or water supplies. This project will identify and characterize opportunistic pathogens in hospital and home premise plumbing systems. The project also considers how the environment as well as an individual’s oral and skin microbiome may contribute to the development of premise plumbing biofilms and investigates potential exposure pathways.
Collaborations
![]()
![]()
![]()
![]()
![]()
![]()
![]()

Claudia Gunsch
Duke, Project Lead

Deverick Anderson
Duke

Sandra Clinton
UNC Charlotte

Lawrence David
Duke
Liesl Jeffers-Francis
N.C. A&T

Joshua Granek
Duke

A-Andrew Jones
Duke

Lingchong You
Duke
RT2-3: Linking physical and chemical properties of premise plumbing and
microbial biofilms
Premise plumbing comprises the complete hot and cold water systems in a building and includes everything from the hot water heater and HVAC to the showers, faucets, sinks, and toilets. These systems are composed of a variety of materials (e.g. copper, PVC, PEX) that result in an environment that varies both temporally and spatially in its physical and chemical properties. This project collects fine scale data on the materials commonly used in premise plumbing and uses it to create a set of known substrates of varying properties that can be used to grow and characterize biofilms under varying environmental conditions.
Collaborations
![]()
![]()
![]()

Sandra Clinton
UNC Charlotte, Project Lead

Claudia Gunsch
Duke

Jacelyn Rice-Boayue
UNC Charlotte

Mark Wiesner
Duke
RT2-4: Identifying determinants of microbial fitness in the built environment
A fundamental challenge in microbiome science is to understand the mechanisms that determine fitness of individual community members in a given habitat. The objective of this project is to establish multiple complementary approaches to defining genes and traits that confer fitness to members of built environment microbiomes. Planned methods include transposon insertion, heterologous expression, and high-throughput sequencing.
Collaborations
![]()
![]()
![]()
![]()

John Rawls
Duke, Project Lead

Joe Brown
UNC Chapel Hill

Nathan Crook
NC State

Glenn Morrison
UNC Chapel Hill

Barbara Turpin
UNC Chapel Hill

Lingchong You
Duke
